Behavioral Neurogenetics
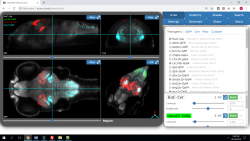 The Challenge: Two major goals in neuroscience are to define neural circuits which select and modulate behavior and to identify genes which contribute to the development of these circuits. Invertebrate species have yielded key insights into behavioral genetics because the nervous system is relatively simple and because their behaviors are stereotyped and genetically specified. Zebrafish larvae have identifiable neurons and a repertoire of genetically determined behaviors. We exploit these advantages of the zebrafish to understand at the cellular and molecular level how neural circuits modulate behavior.
The Challenge: Two major goals in neuroscience are to define neural circuits which select and modulate behavior and to identify genes which contribute to the development of these circuits. Invertebrate species have yielded key insights into behavioral genetics because the nervous system is relatively simple and because their behaviors are stereotyped and genetically specified. Zebrafish larvae have identifiable neurons and a repertoire of genetically determined behaviors. We exploit these advantages of the zebrafish to understand at the cellular and molecular level how neural circuits modulate behavior.
The Solution: We have developed software for zebrafish behavioral and neuroanatomical analysis and a library of enhancer trap lines for manipulating the nervous system. Enhancer trap lines with strong expression in the nervous system are included in our X3D / X3DOM Brain Browser, an atlas of zebrafish transgenic lines. Users can load and compare the expression data from a number of genetic lines at once.
The Contributors: Virginia Tech Visual Computing Group; The Burgess Lab, whichis part of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), located on the NIH campus in Bethesda, just outside of Washington, DC.
Additional References:
- Tabor KM, Marquart GD, Hurt C, Smith TS, Geoca AK, Bhandiwad AA, Subedi A, Sinclair JL, Rose HM, Polys NF and Burgess HA (2019) Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. Elife. doi: 10.7554/eLife.42687.
- Marquart GD, Tabor KM, Horstick EJ, Brown M, Geoca AK, Polys NF, Nogare DD, Burgess HA. High-precision registration between zebrafish brain atlases using symmetric diffeomorphic normalization. GigaScience. 2017 Aug 19;6(8):gix056.
